-Search query
-Search result
Showing all 33 items for (author: rodriguez & uo)

EMDB-27810: 
Cryo-EM structure of chi dynein bound to Lis1
Method: single particle / : Reimer JM, Lahiri I, Leschziner AE

EMDB-27811: 
Symmetry expansion of yeast cytoplasmic dynein-1 bound to Lis1 in the chi conformation.
Method: single particle / : Reimer JM, Lahiri I, Leschziner AE

PDB-8dzz: 
Cryo-EM structure of chi dynein bound to Lis1
Method: single particle / : Reimer JM, Lahiri I, Leschziner AE

PDB-8e00: 
Symmetry expansion of yeast cytoplasmic dynein-1 bound to Lis1 in the chi conformation.
Method: single particle / : Reimer JM, Lahiri I, Leschziner AE

EMDB-40094: 
17b10 fab in complex with full-length SARS-CoV-2 Spike G614 trimer
Method: single particle / : Kwon HJ, Zhang J, Kosikova M, Tang WC, Rodriguez UO, Peng HQ, Meseda CA, Pedro CL, Schmeisser F, Lu JM, Zhou B, Davis CT, Wentworth DE, Chen WH, Shriver MC, Pasetti MF, Weir JP, Chen B, Xie H

EMDB-40095: 
17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer
Method: single particle / : Kwon HJ, Zhang J, Kosikova M, Tang WC, Rodriguez UO, Peng HQ, Meseda CA, Pedro CL, Schmeisser F, Lu JM, Zhou B, Davis CT, Wentworth DE, Chen WH, Shriver MC, Pasetti MF, Weir JP, Chen B, Xie H
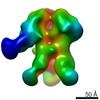
EMDB-9175: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate 224-13
Method: single particle / : Nogal B, Ward AB
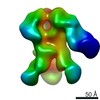
EMDB-9176: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RCn
Method: single particle / : Nogal B, Ward AB
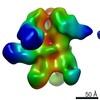
EMDB-9177: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RFr16
Method: single particle / : Nogal B, Ward AB

EMDB-9178: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate ROw16
Method: single particle / : Nogal B, Ward AB

EMDB-9179: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RQq16
Method: single particle / : Nogal B, Ward AB

EMDB-9180: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RRk16
Method: single particle / : Nogal B, Ward AB
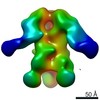
EMDB-9181: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RVh16
Method: single particle / : Nogal B, Ward AB
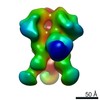
EMDB-9182: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RTh16
Method: single particle / : Nogal B, Ward AB
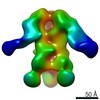
EMDB-9183: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RWh16
Method: single particle / : Nogal B, Ward AB
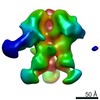
EMDB-9184: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RWr16
Method: single particle / : Nogal B, Ward AB
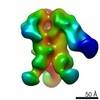
EMDB-9185: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RYm16
Method: single particle / : Nogal B, Ward AB
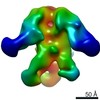
EMDB-9186: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate ROw16
Method: single particle / : Nogal B, Ward AB
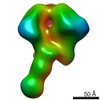
EMDB-0569: 
HIV-1 vaccine elicited polyclonal 5N6 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB
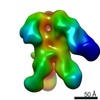
EMDB-0570: 
HIV-1 vaccine elicited polyclonal 99-13 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0571: 
HIV-1 vaccine elicited polyclonal 145-11 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB
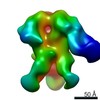
EMDB-0572: 
HIV-1 vaccine elicited polyclonal BM57 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0573: 
HIV-1 vaccine elicited polyclonal BO77 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0574: 
HIV-1 vaccine elicited polyclonal BO77 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB
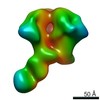
EMDB-0575: 
HIV-1 vaccine elicited polyclonal RAa14 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0576: 
HIV-1 vaccine elicited polyclonal RAv16 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0577: 
HIV-1 vaccine elicited polyclonal RAv16 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB
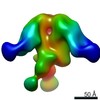
EMDB-0578: 
HIV-1 vaccine elicited polyclonal REj15 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0579: 
HIV-1 vaccine elicited polyclonal REv16 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0580: 
HIV-1 vaccine elicited polyclonal RHw16 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0581: 
HIV-1 vaccine elicited polyclonal RMI15 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0582: 
HIV-1 vaccine elicited polyclonal RWo16 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB
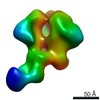
EMDB-9138: 
HIV-1 vaccine elicited Fab BDA1 in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Cottrell CA, Ward AB
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
